

Our approach uses a technique called Raman spectroscopy to identify viruses by shining a light on a disposable cartridge that collects samples from oral cotton swabs



The Research Brief is a short take about interesting academic work.
The big idea
The most common type of test for the new coronavirus takes several hours and is uncomfortable; samples are obtained by sliding a swab into the nose or throat.

I am collaborating with other scientists, including Yin-Ting Yeh at Penn State, Elodie Ghedin at New York University, Shengxi Huang at Penn State and Sharon X Huang at Penn State, on a diagnostic tool to rapidly trap and identify viruses using a laser beam and a detector. The team includes myself, a physicist, as well virologists, engineers, chemists and data scientists.
How we do the work
Our approach uses a technique called Raman spectroscopy to identify viruses by shining a light on a disposable cartridge that collects samples from oral cotton swabs or a person blowing through the device. Once a sample is collected, a spectrometer measures the interatomic vibrations that result from shining the light on the collected viruses. Each virus has its own signature vibrations, which act as a sort of optical fingerprint that can distinguish the coronavirus from, for example, the virus that causes influenza.
We could capture viruses from patients’ saliva taken with a swab or by a person blowing through a device, called a microfluidic cartridge. The air and liquid pass an array of carbon nanotubes, cylinder-shape molecules used in different materials.
The diameters of the nanotubes are microscopic, between 10-60 nanometers. Because they are smaller than microbes — flu viruses range from 90-120 nanometers in diameter and coronaviruses range from 125-150nm in diameter — the pathogens collect on the carbon nanotubes. Once trapped by passing through the carbon nanotubes, the viruses can be optically identified by shining a laser on the sample. Shining the light on the carbon nanotubes and pathogens creates a distinctive optical fingerprint, or ‘Raman peaks’.
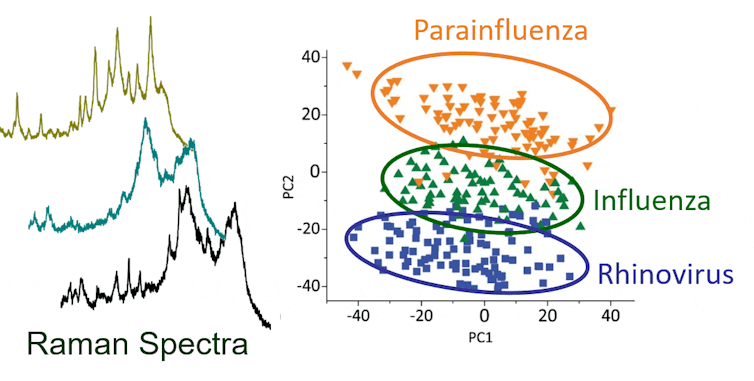
After the laser shines on the trapped sample, machine learning algorithms identify the signature spectrum of the virus that results from the light that bounces off the virus particles. With the assistance of machine learning, the identification takes less than two minutes with an accuracy rate of up to 70-90 per cent, comparable to state-of-the-art microbiology techniques.
Why it matters
Right now, the rapid and accurate detection of the novel coronavirus is of paramount importance. While Raman spectroscopy has the potential to be enormously helpful in identifying this virus, doctors can also use this technique to test for other illnesses, such as influenza. By identifying the virus easily, quickly and at the point of contact, Raman spectroscopy could significantly halt disease spread.
Compare that to our current methods of analysing samples: A process that is relatively slow, tedious, labor intensive and requires extensive scrutiny at laboratories. Early and rapid detection with this new device has the potential to save hundreds of thousands of lives every year.
What other work is being done
For the identification of viruses, existing technologies do provide relatively sensitive detection. However, they take several hours and sometime days depending on the quality of the sample collected because low virus concentrations are very difficult to process and results in false negatives.
Unfortunately, both immune- and molecular-based methods, including enzyme-linked immunosorbent assay (ELISA) and polymerase chain reaction (PCR), require prior knowledge of the strains. Another technique known as deep sequencing is another promising new approach, but obtaining sufficient viral reads for it to work well depends on the quality of the sample and its preparation. Processing steps involve incorporating different benchtop equipment, reagents and technical expertise. This Raman technique has been recently developed to identify different bacteria, thus demonstrating the technique is indeed novel and viable.
What’s next
We are applying for federal funds to demonstrate that this technology works for SARS-CoV-2, the virus that causes COVID-19, and then build reliable prototypes that can be scaled up for mass production and field deployment. We are also talking with several manufacturers and exploring ways to move the technology forward to help in the current crisis.
We have been successful in capturing human respiratory viruses from clinical samples using this technique. Eventually, we foresee this technology becoming available to anyone visiting their family doctor. Within two minutes, a person would know whether you have a respiratory virus by comparing the result of the spectroscopy test with other results in a database. In the future, this technology could be at hospitals, airports and inside commercial aircraft to avoid outbreaks. And the captured viruses, still viable, can be replicated to develop a vaccine.
[You’re smart and curious about the world. So are The Conversation’s authors and editors. You can get our highlights each weekend.]![]()
Mauricio Terrones, Professor of Physics, Pennsylvania State University
This article is republished from The Conversation under a Creative Commons license. Read the original article.
We are a voice to you; you have been a support to us. Together we build journalism that is independent, credible and fearless. You can further help us by making a donation. This will mean a lot for our ability to bring you news, perspectives and analysis from the ground so that we can make change together.

Comments are moderated and will be published only after the site moderator’s approval. Please use a genuine email ID and provide your name. Selected comments may also be used in the ‘Letters’ section of the Down To Earth print edition.